%matplotlib inline
from preamble import *
Representing Data and Engineering Features¶
- Categorical Variables
- One-Hot-Encoding (Dummy Variables)
- Table of Contents
- Numbers Can Encode Categoricals
- Binning, Discretization, Linear Models, and Trees
- Interactions and Polynomials
- Univariate Nonlinear Transformations
- Automatic Feature Selection
- Univariate Statistics
- Model-Based Feature Selection
- Iterative Feature Selection
- Utilizing Expert Knowledge
- Summary and Outlook
0. Data and variables¶
Most data can be categorized into 4 basic types from a Machine Learning perspective: numerical data, categorical data, time series data, and text.
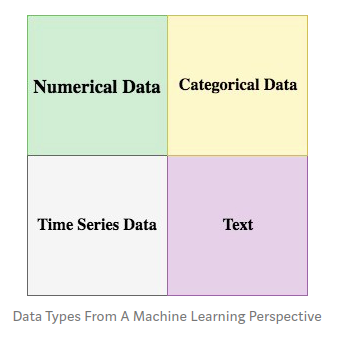
Numerical Data¶
Numerical data is any data where data points are exact numbers.
Categorical Data¶
Categorical data represents characteristics, such as a hockey player’s position, team, hometown .
Time Series Data¶
Time series data is a sequence of numbers collected at regular intervals over some period of time.
Text¶
Text data is basically just words.
Variables¶
There is 2 main types of variables:
- Numerical variables or quantitative variables:
- Descrete
- Continuous
- Categorical variables or qualitative variables:
- Nominal
- Ordinal
- Binary
1. Categorical Variables¶
One-Hot-Encoding (Dummy variables)¶
import os
# The file has no headers naming the columns, so we pass header=None
# and provide the column names explicitly in "names"
adult_path = os.path.join(mglearn.datasets.DATA_PATH, "adult.data")
data = pd.read_csv(
adult_path, header=None, index_col=False,
names=['age', 'workclass', 'fnlwgt', 'education', 'education-num',
'marital-status', 'occupation', 'relationship', 'race', 'gender',
'capital-gain', 'capital-loss', 'hours-per-week', 'native-country',
'income'])
# For illustration purposes, we only select some of the columns
data = data[['age', 'workclass', 'education', 'gender', 'hours-per-week',
'occupation', 'income']]
# IPython.display allows nice output formatting within the Jupyter notebook
display(data.head())
In this dataset, age and hours-per-week are continuous features, which we know how to treat. The workclass, education, sex, and occupation features are categorical => denote a qualitative property
Checking string-encoded categorical data¶
By far the most common way to represent categorical variables is using the one-hot encoding or one-out-of-N encoding, also known as dummy variables.
The idea behind dummy variables is to replace a categorical variable with one or more new features that can have the values 0 and 1.
After reading a dataset like this, it is often good to first check if a column actually contains meaningful categorical data.
print(data.gender.value_counts())
We can see that there are exactly two values for gender in this dataset, Male and Female, meaning the data is already in a good format to be represented using onehot-encoding. In a real application, you should look at all columns and check their values.
print("Original features:\n", list(data.columns), "\n")
data_dummies = pd.get_dummies(data)
print("Features after get_dummies:\n", list(data_dummies.columns))
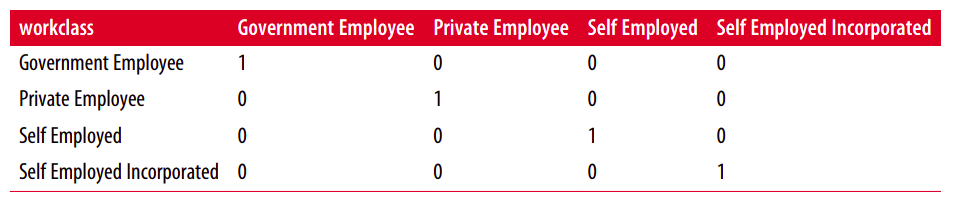
display(data_dummies.head())
features = data_dummies.loc[:, 'age':'occupation_ Transport-moving']
# Extract NumPy arrays
X = features.values
y = data_dummies['income_ >50K'].values
print("X.shape: {} y.shape: {}".format(X.shape, y.shape))
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0)
logreg = LogisticRegression()
logreg.fit(X_train, y_train)
print("Test score: {:.2f}".format(logreg.score(X_test, y_test)))
Numbers Can Encode Categoricals¶
# create a DataFrame with an integer feature and a categorical string feature
demo_df = pd.DataFrame({'Integer Feature': [0, 1, 2, 1],
'Categorical Feature': ['socks', 'fox', 'socks', 'box']})
display(demo_df)
display(pd.get_dummies(demo_df))
demo_df['Integer Feature'] = demo_df['Integer Feature'].astype(str)
display(pd.get_dummies(demo_df, columns=['Integer Feature', 'Categorical Feature']))
In statistics, it is common to encode a categorical feature with k different possible values into k–1 features (the last one is represented as all zeros).
Note:
- Categorical features are often encoded using integers. That they are numbers doesn’t mean that they should necessarily be treated as continuous features.
- It is not always clear whether an integer feature should be treated as continuous or discrete (and one-hotencoded).
- If there is no ordering between the semantics that are encoded (like in the workclass example), the feature must be treated as discrete.
OneHotEncoder and ColumnTransformer: Categorical Variables with scikit-learn¶
from sklearn.preprocessing import OneHotEncoder
# Setting sparse=False means OneHotEncode will return a numpy array, not a sparse matrix
ohe = OneHotEncoder(sparse=False)
print(ohe.fit_transform(demo_df))
print(ohe.get_feature_names())
display(data.head())
from sklearn.compose import ColumnTransformer
from sklearn.preprocessing import StandardScaler
ct = ColumnTransformer(
[("scaling", StandardScaler(), ['age', 'hours-per-week']),
("onehot", OneHotEncoder(sparse=False), ['workclass', 'education', 'gender', 'occupation'])])
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
# get all columns apart from income for the features
data_features = data.drop("income", axis=1)
# split dataframe and income
X_train, X_test, y_train, y_test = train_test_split(
data_features, data.income, random_state=0)
ct.fit(X_train)
X_train_trans = ct.transform(X_train)
print(X_train_trans.shape)
logreg = LogisticRegression()
logreg.fit(X_train_trans, y_train)
X_test_trans = ct.transform(X_test)
print("Test score: {:.2f}".format(logreg.score(X_test_trans, y_test)))
ct.named_transformers_.onehot
Convenient ColumnTransformer creation with make_columntransformer¶
from sklearn.compose import make_column_transformer
ct = make_column_transformer(
(['age', 'hours-per-week'], StandardScaler()),
(['workclass', 'education', 'gender', 'occupation'], OneHotEncoder(sparse=False)))
2. Binning(Discretization) Linear Models, and Trees¶
The best way to represent data depends not only on the semantics of the data, but also on the kind of model you are using.
- Linear models and tree-based models (such as decision trees, gradient boosted trees, and random forests), two large and very commonly used families, have very different properties when it comes to how they work with different feature representations.
- Linear models can only model linear relationships, which are lines in the case of a single feature.
- The decision tree can build a much more complex model of the data. However, this is strongly dependent on the representation of the data.
Binning¶
One way to make linear models more powerful on continuous data is to use binning (also known as discretization) of the feature to split it up into multiple features
Binning can be applied on both categorical and numerical data:
Numerical Binning Example¶
Value Bin
0-30 -> Low
31-70 -> Mid
71-100 -> High
Categorical Binning Example¶
Value Bin
Spain -> Europe
Italy -> Europe
Chile -> South America
Brazil -> South America
The main motivation of binning is to make the model more robust and prevent overfitting, however, it has a cost to the performance.
Every time we bin something, we sacrifice information and make your data more regularized.
from sklearn.linear_model import LinearRegression
from sklearn.tree import DecisionTreeRegressor
X, y = mglearn.datasets.make_wave(n_samples=120)
line = np.linspace(-3, 3, 1000, endpoint=False).reshape(-1, 1)
reg = DecisionTreeRegressor(min_samples_leaf=3).fit(X, y)
plt.plot(line, reg.predict(line), label="decision tree")
reg = LinearRegression().fit(X, y)
plt.plot(line, reg.predict(line), label="linear regression")
plt.plot(X[:, 0], y, 'o', c='k')
plt.ylabel("Regression output")
plt.xlabel("Input feature")
plt.legend(loc="best")
A partition of the input range for the feature (in this case, the numbers from –3 to 3) into a fixed number of bins—say, 1000
from sklearn.preprocessing import KBinsDiscretizer
kb = KBinsDiscretizer(n_bins=10, strategy='uniform')
kb.fit(X)
print("bin edges: \n", kb.bin_edges_)
X_binned = kb.transform(X)
X_binned
print(X[:10])
X_binned.toarray()[:10]
kb = KBinsDiscretizer(n_bins=10, strategy='uniform', encode='onehot-dense')
kb.fit(X)
X_binned = kb.transform(X)
line_binned = kb.transform(line)
reg = LinearRegression().fit(X_binned, y)
plt.plot(line, reg.predict(line_binned), label='linear regression binned')
reg = DecisionTreeRegressor(min_samples_split=3).fit(X_binned, y)
plt.plot(line, reg.predict(line_binned), label='decision tree binned')
plt.plot(X[:, 0], y, 'o', c='k')
plt.vlines(kb.bin_edges_[0], -3, 3, linewidth=1, alpha=.2)
plt.legend(loc="best")
plt.ylabel("Regression output")
plt.xlabel("Input feature")
The linear line and tree line are exactly on top of each other, meaning the linear regression model and the decision tree make exactly the same predictions.
- For each bin, they predict a constant value. As features are constant within each bin, any model must predict the same value for all points within a bin.
- Comparing what the models learned before binning the features and after, we see that the linear model became much more flexible, because it now has a different value for each bin, while the decision tree model got much less flexible.
- Binning features generally has no beneficial effect for tree-based models, as these models can learn to split up the data anywhere. In a sense, that means decision trees can learn whatever binning is most useful for predicting on this data.
Additionally, decision trees look at multiple features at once, while binning is usually done on a per-feature basis. However, the linear model benefited greatly in expressiveness from the transformation of the data.
Interactions and Polynomials¶
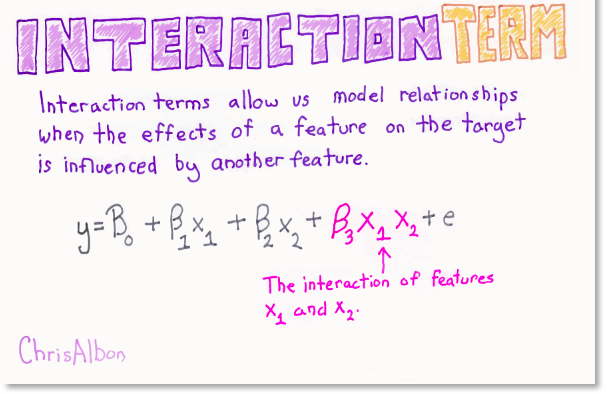
Another way to enrich a feature representation, particularly for linear models, is adding interaction features and polynomial features of the original data.
This kind of feature engineering is often used in statistical modeling, but it’s also common in many practical machine learning applications.
Interaction effects can be account for by including a new feature comprising the product of corresponding values from the interacting features
Polynomials feature is a technique to convert the original features into their higher order terms.
- To overcome under-fitting, we need to increase the complexity of the model, but we do not have abilities to add more another feature (have no time to collect more features, how to reuse current features)
X_combined = np.hstack([X, X_binned])
print(X_combined.shape)
The linear model learned a constant value for each bin in the wave dataset.
- Linear models can learn not only offsets, but also slopes. => One way to add a slope to the linear model on the binned data is to add the original feature (the x-axis in the plot) back in.
reg = LinearRegression().fit(X_combined, y)
line_combined = np.hstack([line, line_binned])
plt.plot(line, reg.predict(line_combined), label='linear regression combined')
plt.vlines(kb.bin_edges_[0], -3, 3, linewidth=1, alpha=.2)
plt.legend(loc="best")
plt.ylabel("Regression output")
plt.xlabel("Input feature")
plt.plot(X[:, 0], y, 'o', c='k')
The model learned an offset for each bin, together with a slope. The learned slope is downward, and shared across all the bins—there is a single x-axis feature, which has a single slope
X_product = np.hstack([X_binned, X * X_binned])
print(X_product.shape)
We would rather have a separate slope for each bin!
- We can achieve this by adding an interaction or product feature that indicates which bin a data point is in and where it lies on the x-axis.
- This feature is a product of the bin indicator and the original feature.
reg = LinearRegression().fit(X_product, y)
line_product = np.hstack([line_binned, line * line_binned])
plt.plot(line, reg.predict(line_product), label='linear regression product')
plt.vlines(kb.bin_edges_[0], -3, 3, linewidth=1, alpha=.2)
plt.plot(X[:, 0], y, 'o', c='k')
plt.ylabel("Regression output")
plt.xlabel("Input feature")
plt.legend(loc="best")
As you can see, now each bin has its own offset and slope in this model. Using binning is one way to expand a continuous feature.
Another one is to use polynomials of the original features.
from sklearn.preprocessing import PolynomialFeatures
# include polynomials up to x ** 10:
# the default "include_bias=True" adds a feature that's constantly 1
poly = PolynomialFeatures(degree=10, include_bias=False)
poly.fit(X)
X_poly = poly.transform(X)
For a given feature x, we might want to consider x 2, x 3, x ** 4, and so on.
print("X_poly.shape: {}".format(X_poly.shape))
print("Entries of X:\n{}".format(X[:5]))
print("Entries of X_poly:\n{}".format(X_poly[:5]))
print("Polynomial feature names:\n{}".format(poly.get_feature_names()))
Using polynomial features together with a linear regression model yields the classical model of polynomial regression.
As you can see, polynomial features yield a very smooth fit on this one-dimensional data.
reg = LinearRegression().fit(X_poly, y)
line_poly = poly.transform(line)
plt.plot(line, reg.predict(line_poly), label='polynomial linear regression')
plt.plot(X[:, 0], y, 'o', c='k')
plt.ylabel("Regression output")
plt.xlabel("Input feature")
plt.legend(loc="best")
from sklearn.svm import SVR
for gamma in [1, 10]:
svr = SVR(gamma=gamma).fit(X, y)
plt.plot(line, svr.predict(line), label='SVR gamma={}'.format(gamma))
plt.plot(X[:, 0], y, 'o', c='k')
plt.ylabel("Regression output")
plt.xlabel("Input feature")
plt.legend(loc="best")
from sklearn.datasets import load_boston
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler
boston = load_boston()
X_train, X_test, y_train, y_test = train_test_split(
boston.data, boston.target, random_state=0)
# rescale data
scaler = MinMaxScaler()
X_train_scaled = scaler.fit_transform(X_train)
X_test_scaled = scaler.transform(X_test)
poly = PolynomialFeatures(degree=2).fit(X_train_scaled)
X_train_poly = poly.transform(X_train_scaled)
X_test_poly = poly.transform(X_test_scaled)
print("X_train.shape: {}".format(X_train.shape))
print("X_train_poly.shape: {}".format(X_train_poly.shape))
print("Polynomial feature names:\n{}".format(poly.get_feature_names()))
from sklearn.linear_model import Ridge
ridge = Ridge().fit(X_train_scaled, y_train)
print("Score without interactions: {:.3f}".format(
ridge.score(X_test_scaled, y_test)))
ridge = Ridge().fit(X_train_poly, y_train)
print("Score with interactions: {:.3f}".format(
ridge.score(X_test_poly, y_test)))
from sklearn.ensemble import RandomForestRegressor
rf = RandomForestRegressor(n_estimators=100).fit(X_train_scaled, y_train)
print("Score without interactions: {:.3f}".format(
rf.score(X_test_scaled, y_test)))
rf = RandomForestRegressor(n_estimators=100).fit(X_train_poly, y_train)
print("Score with interactions: {:.3f}".format(rf.score(X_test_poly, y_test)))
Univariate Nonlinear Transformations¶
- A nonlinear transformation changes (increases or decreases) linear relationships between variables and, thus, changes the correlation between variables. Examples of nonlinear transformation of variable x would be taking the square root x or the reciprocal of x.
There are other transformations that often prove useful for transforming certain features: in particular, applying mathematical functions like log, exp, or sin.
If this transformation increases the linearity of the relationship between the variables, it allows the analyst to use linear regression techniques appropriately with nonlinear data.
rnd = np.random.RandomState(0)
X_org = rnd.normal(size=(1000, 3))
w = rnd.normal(size=3)
X = rnd.poisson(10 * np.exp(X_org))
y = np.dot(X_org, w)
print("Number of feature appearances:\n{}".format(np.bincount(X[:, 0])))
bins = np.bincount(X[:, 0])
plt.bar(range(len(bins)), bins, color='grey')
plt.ylabel("Number of appearances")
plt.xlabel("Value")
from sklearn.linear_model import Ridge
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0)
score = Ridge().fit(X_train, y_train).score(X_test, y_test)
print("Test score: {:.3f}".format(score))
X_train_log = np.log(X_train + 1)
X_test_log = np.log(X_test + 1)
plt.hist(X_train_log[:, 0], bins=25, color='gray')
plt.ylabel("Number of appearances")
plt.xlabel("Value")
score = Ridge().fit(X_train_log, y_train).score(X_test_log, y_test)
print("Test score: {:.3f}".format(score))
Note:
- Binning, polynomials, and interactions can have a huge influence on how models perform on a given dataset.
- Tree-based models, on the other hand, are often able to discover important interactions them selves, and don’t require transforming the data explicitly most of the time.
- SVMs, nearest neighbors, and neural networks, might sometimes benefit from using binning, interactions, or polynomials, but the implications there are usually much less clear than in the case of linear models.
3. Automatic Feature Selection¶
- Adding more features makes all models more complex, and so increases the chance of overfitting.
- Adding new features, or with high-dimensional datasets in general, it can be a good idea to reduce the number of features to only the most useful ones,and discard the rest.
There are three basic strategies:
- univariate statistics
- model-based selection
- iterative selection.
All of these methods are supervised methods, meaning they need the target for fitting the model. This means we need to split the data into training and test sets, and fit the feature selection only on the training part of the data.
3.1 Univariate statistics¶
In univariate statistics, we compute whether there is a statistically significant relationship between each feature and the target. Then the features that are related with the highest confidence are selected.
In the case of classification, this is also known as analysis of variance (ANOVA). A key property of these tests is that they are univariate, meaning that they only consider each feature individually. Consequently, a fea‐ ture will be discarded if it is only informative when combined with another feature.
- Univariate tests are often very fast to compute, and don’t require building a model.
- On the other hand, they are completely independent of the model that you might want to apply after the feature selection.
from sklearn.datasets import load_breast_cancer
from sklearn.feature_selection import SelectPercentile
from sklearn.model_selection import train_test_split
cancer = load_breast_cancer()
# get deterministic random numbers
rng = np.random.RandomState(42)
noise = rng.normal(size=(len(cancer.data), 50))
# add noise features to the data
# the first 30 features are from the dataset, the next 50 are noise
X_w_noise = np.hstack([cancer.data, noise])
X_train, X_test, y_train, y_test = train_test_split(
X_w_noise, cancer.target, random_state=0, test_size=.5)
# use f_classif (the default) and SelectPercentile to select 50% of features
select = SelectPercentile(percentile=50)
select.fit(X_train, y_train)
# transform training set
X_train_selected = select.transform(X_train)
print("X_train.shape: {}".format(X_train.shape))
print("X_train_selected.shape: {}".format(X_train_selected.shape))
Note:
To use univariate feature selection, we need to choose a test, usually either f_classif (the default) for classification or f_regression for regression, and a method to discard features based on the p-values determined in the test.
All methods for discarding parameters use a threshold to discard all features with too high a p-value (which means they are unlikely to be related to the target).
The methods differ in how they compute this threshold, with the simplest ones being SelectKBest, which selects a fixed number k of features, and SelectPercentile, which selects a fixed percentage of features.
We expect the feature selection to be able to identify the features that are noninformative and remove them
mask = select.get_support()
print(mask)
# visualize the mask. black is True, white is False
plt.matshow(mask.reshape(1, -1), cmap='gray_r')
plt.xlabel("Sample index")
plt.yticks(())
from sklearn.linear_model import LogisticRegression
# transform test data
X_test_selected = select.transform(X_test)
lr = LogisticRegression()
lr.fit(X_train, y_train)
print("Score with all features: {:.3f}".format(lr.score(X_test, y_test)))
lr.fit(X_train_selected, y_train)
print("Score with only selected features: {:.3f}".format(
lr.score(X_test_selected, y_test)))
Univariate feature selection can still be very helpful, though, if there is such a large number of features that building a model on them is infeasible, or if you suspect that many features are completely uninformative
3.2 Model-based Feature Selection¶
Model-based feature selection uses a supervised machine learning model to judge the importance of each feature, and keeps only the most important ones.
- The supervised model that is used for feature selection doesn’t need to be the same model that is used for the final supervised modeling.
- The feature selection model needs to provide some measure of importance for each feature, so that they can be ranked by this measure.
Decision trees and decision tree–based models provide a featureimportances attribute, which directly encodes the importance of each feature.
Linear models have coefficients, which can also be used to capture feature importances by considering the absolute values
from sklearn.feature_selection import SelectFromModel
from sklearn.ensemble import RandomForestClassifier
select = SelectFromModel(
RandomForestClassifier(n_estimators=100, random_state=42),
threshold="median")
select.fit(X_train, y_train)
X_train_l1 = select.transform(X_train)
print("X_train.shape: {}".format(X_train.shape))
print("X_train_l1.shape: {}".format(X_train_l1.shape))
mask = select.get_support()
# visualize the mask. black is True, white is False
plt.matshow(mask.reshape(1, -1), cmap='gray_r')
plt.xlabel("Sample index")
plt.yticks(())
X_test_l1 = select.transform(X_test)
score = LogisticRegression().fit(X_train_l1, y_train).score(X_test_l1, y_test)
print("Test score: {:.3f}".format(score))
3.3 Iterative feature selection¶
In univariate testing we used no model, while in model-based selection we used a single model to select features.
In iterative feature selection, a series of models are built, with varying numbers of features
There are two basic methods:
- Starting with no features and adding features one by one until some stopping criterion is reached,
- Starting with all features and removing features one by one until some stopping criterion is reached.
Because a series of models are built, these methods are much more computationally expensive than the methods we discussed previously.
One particular method of this kind is recursive feature elimination (RFE),
- Starts with all fea‐ tures, builds a model, and discards the least important feature according to the model.
- Then a new model is built using all but the discarded feature, and so on until only a prespecified number of features are left.
For this to work, the model used for selection needs to provide some way to determine feature importance, as was the case for the model-based selection.
from sklearn.feature_selection import RFE
select = RFE(RandomForestClassifier(n_estimators=100, random_state=42),
n_features_to_select=40)
select.fit(X_train, y_train)
# visualize the selected features:
mask = select.get_support()
plt.matshow(mask.reshape(1, -1), cmap='gray_r')
plt.xlabel("Sample index")
plt.yticks(())
The feature selection got better compared to the univariate and model-based selection, but one feature was still missed. Running above code also takes significantly longer than that for the model-based selection, because a random forest model is trained 40 times, once for each feature that is dropped.
X_train_rfe = select.transform(X_train)
X_test_rfe = select.transform(X_test)
score = LogisticRegression().fit(X_train_rfe, y_train).score(X_test_rfe, y_test)
print("Test score: {:.3f}".format(score))
print("Test score: {:.3f}".format(select.score(X_test, y_test)))
If you are unsure when selecting what to use as input to your machine learning algorithms, automatic feature selection can be quite helpful. It is also great for reducing the amount of features needed for example, to speed up prediction or to allow for more interpretable models. In most real-world cases, applying feature selection is unlikely to provide large gains in performance.
4. Utilizing Expert Knowledge¶
Feature engineering is often an important place to use expert knowledge for a particular application. While the purpose of machine learning in many cases is to avoid having to create a set of expert-designed rules, that doesn’t mean that prior knowledge of the application or domain should be discarded.
Often, domain experts can help in identifying useful features that are much more informative than the initial representation of the data.
Prior knowledge about the nature of the task can be encoded in the features to aid a machine learning algorithm.
citibike = mglearn.datasets.load_citibike()
print("Citi Bike data:\n{}".format(citibike.head()))
plt.figure(figsize=(10, 3))
xticks = pd.date_range(start=citibike.index.min(), end=citibike.index.max(),
freq='D')
plt.xticks(xticks, xticks.strftime("%a %m-%d"), rotation=90, ha="left")
plt.plot(citibike, linewidth=1)
plt.xlabel("Date")
plt.ylabel("Rentals")
# extract the target values (number of rentals)
y = citibike.values
# convert to POSIX time by dividing by 10**9
X = citibike.index.astype("int64").values.reshape(-1, 1) // 10**9
# use the first 184 data points for training, the rest for testing
n_train = 184
# function to evaluate and plot a regressor on a given feature set
def eval_on_features(features, target, regressor):
# split the given features into a training and a test set
X_train, X_test = features[:n_train], features[n_train:]
# also split the target array
y_train, y_test = target[:n_train], target[n_train:]
regressor.fit(X_train, y_train)
print("Test-set R^2: {:.2f}".format(regressor.score(X_test, y_test)))
y_pred = regressor.predict(X_test)
y_pred_train = regressor.predict(X_train)
plt.figure(figsize=(10, 3))
plt.xticks(range(0, len(X), 8), xticks.strftime("%a %m-%d"), rotation=90,
ha="left")
plt.plot(range(n_train), y_train, label="train")
plt.plot(range(n_train, len(y_test) + n_train), y_test, '-', label="test")
plt.plot(range(n_train), y_pred_train, '--', label="prediction train")
plt.plot(range(n_train, len(y_test) + n_train), y_pred, '--',
label="prediction test")
plt.legend(loc=(1.01, 0))
plt.xlabel("Date")
plt.ylabel("Rentals")
from sklearn.ensemble import RandomForestRegressor
regressor = RandomForestRegressor(n_estimators=100, random_state=0)
eval_on_features(X, y, regressor)
X_hour = citibike.index.hour.values.reshape(-1, 1)
eval_on_features(X_hour, y, regressor)
X_hour_week = np.hstack([citibike.index.dayofweek.values.reshape(-1, 1),
citibike.index.hour.values.reshape(-1, 1)])
eval_on_features(X_hour_week, y, regressor)
from sklearn.linear_model import LinearRegression
eval_on_features(X_hour_week, y, LinearRegression())
enc = OneHotEncoder()
X_hour_week_onehot = enc.fit_transform(X_hour_week).toarray()
eval_on_features(X_hour_week_onehot, y, Ridge())
poly_transformer = PolynomialFeatures(degree=2, interaction_only=True,
include_bias=False)
X_hour_week_onehot_poly = poly_transformer.fit_transform(X_hour_week_onehot)
lr = Ridge()
eval_on_features(X_hour_week_onehot_poly, y, lr)
hour = ["%02d:00" % i for i in range(0, 24, 3)]
day = ["Mon", "Tue", "Wed", "Thu", "Fri", "Sat", "Sun"]
features = day + hour
features_poly = poly_transformer.get_feature_names(features)
features_nonzero = np.array(features_poly)[lr.coef_ != 0]
coef_nonzero = lr.coef_[lr.coef_ != 0]
plt.figure(figsize=(15, 2))
plt.plot(coef_nonzero, 'o')
plt.xticks(np.arange(len(coef_nonzero)), features_nonzero, rotation=90)
plt.xlabel("Feature name")
plt.ylabel("Feature magnitude")
Summary and Outlook¶
In this chapter, we discussed how to deal with different data types (in particular, with categorical variables). We emphasized the importance of representing data in a way that is suitable for the machine learning algorithm—for example, by one-hotencoding categorical variables.
We also discussed the importance of engineering new features, and the possibility of utilizing expert knowledge in creating derived features from your data.
In particular, linear models might benefit greatly from generating new features via binning and adding polynomials and interactions, while more complex, nonlinear models like random forests and SVMs might be able to learn more complex tasks without explicitly expanding the feature space.
In practice, the features that are used (and the match between features and method) is often the most important piece in making a machine learning approach work well.
Comments !